First generation Sanger sequencing
First-generation Sanger sequencing is a technique to detect nucleic acid sequence based on dideoxy chain-termination method developed by Frederick Sanger. Ever since the first genetic analyzer in 1986, Sanger sequencing has gone through several technological innovations.
Multi-capillary fluorescence labeling capillary electrophoresis sequencing has been popularized nowadays. Its principle is: four fluorescently labeled ddNTPs are incorporated into dNTPs for PCR amplification; the random incorporation of ddNTP on every site of the PCR products and the lack of OH group cause DNA polymerase to cease the extension of DNA; After amplification, thousands of fragments of different sizes are obtained, undergo capillary electrophoresis and are fluorescently detected to obtain the sequence of the target fragment.
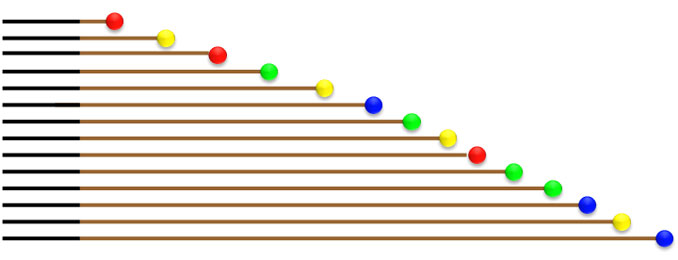
Fig. 1 Diagram of Capillary Electrophoresis for PCR Products
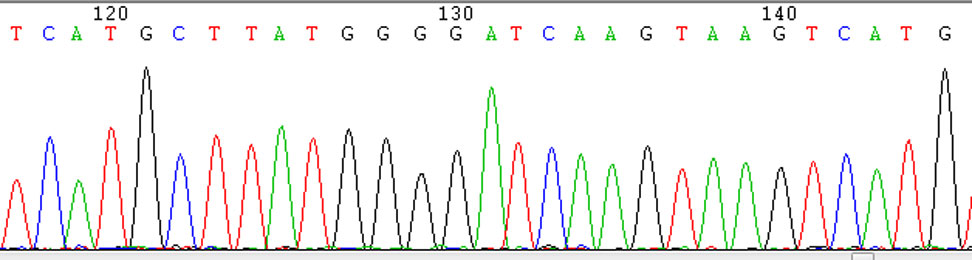
Fig. 2 Results of Sanger Sequencing
Sanger sequencing can directly obtain sequence information of nucleic acid and is considered as the gold standard for attaining nucleic acid sequence information nowadays. In molecular biology researches, de novo sequencing and re-sequencing of plasmids and amplicons are the basis for many researches. In clinical medicine, Sanger sequencing is widely used in molecular diagnosis fields such as pathogen detection, pharmacogenomics and oncogene detection, etc.
By now, we have introduced 3500 DX(8-capillary), 3130(4-capillary) and 3130xl(16-capillary) sequencing platforms from Applied Systems. Based on these sequencing platforms, we are carrying out R&D of several clinical molecular diagnostics reagents including pharmacogenomics, oncogene detection, etc., which will be gradually extended to basic R&D, clinical researches, etc. to lay an important foundation for improving technical capacities across all platforms.




